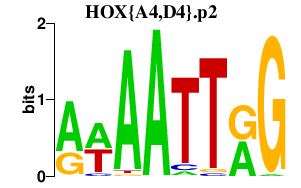
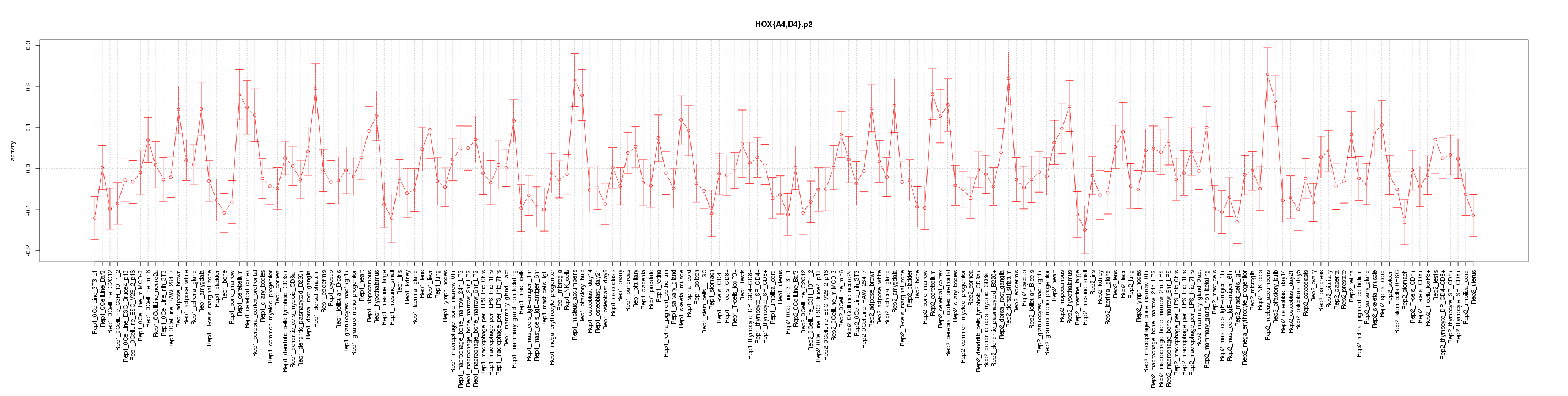
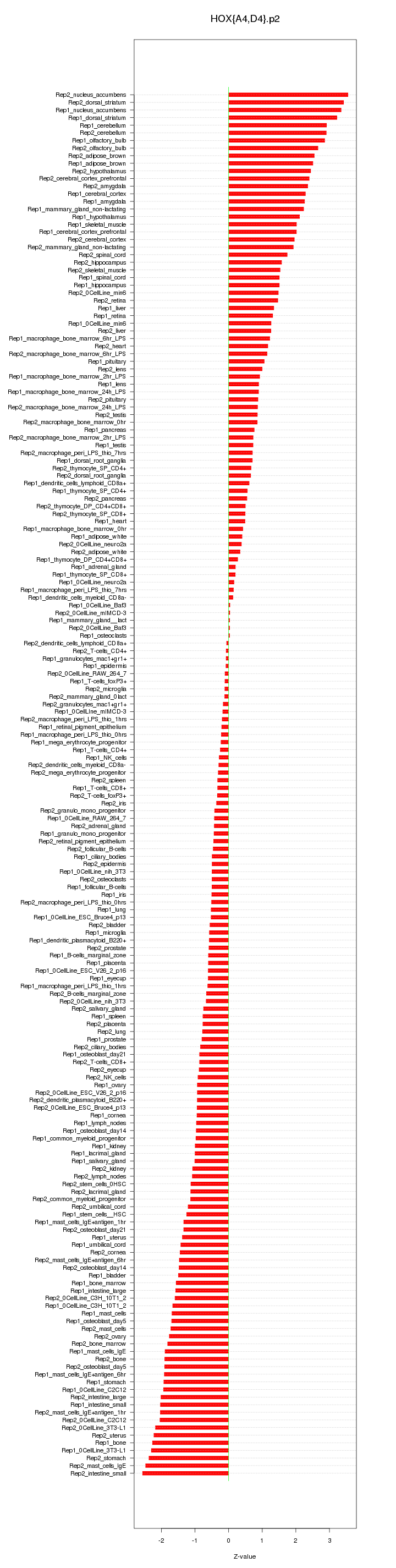
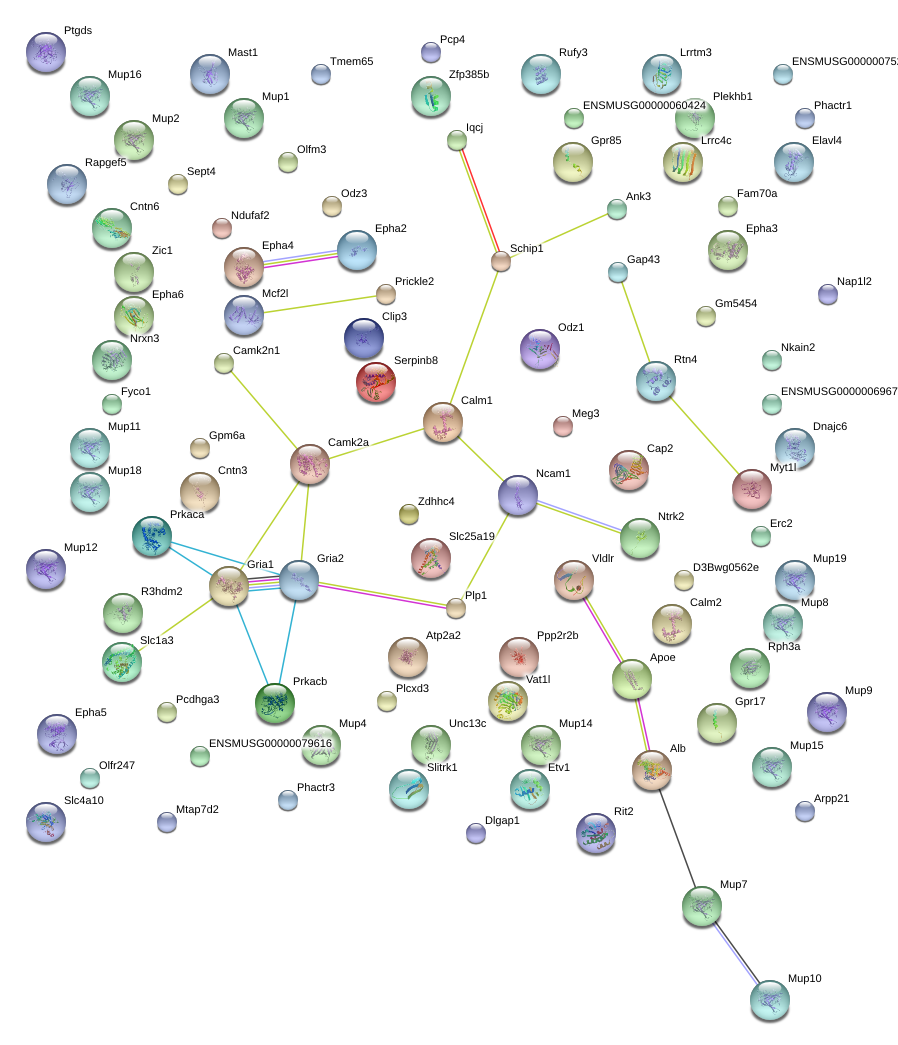
|
chr10_+_69487700
|
31.171
|
|
Ank3
|
ankyrin 3, epithelial
|
|
chr5_-_122906470
|
28.190
|
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chrX_+_133357268
|
24.343
|
NM_011123
|
Plp1
|
proteolipid protein (myelin) 1
|
|
chr17_+_71168939
|
23.795
|
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr11_+_56825307
|
21.677
|
|
Gria1
|
glutamate receptor, ionotropic, AMPA1 (alpha 1)
|
|
chr16_+_7069971
|
21.595
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr11_+_56825258
|
21.243
|
|
Gria1
|
glutamate receptor, ionotropic, AMPA1 (alpha 1)
|
|
chr5_-_122907002
|
20.888
|
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr7_-_107810868
|
20.027
|
NM_001163182
NM_013746
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr12_+_110784600
|
19.877
|
|
Meg3
|
maternally expressed 3
|
|
chr11_+_56825192
|
19.766
|
NM_001113325
NM_008165
|
Gria1
|
glutamate receptor, ionotropic, AMPA1 (alpha 1)
|
|
chr16_+_7069927
|
19.580
|
NM_183188
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr12_+_91570569
|
19.105
|
|
Nrxn3
|
neurexin III
|
|
chr12_+_30219769
|
18.605
|
NM_001093776
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr2_+_177910053
|
18.451
|
NM_001177791
|
Phactr3
|
phosphatase and actin regulator 3
|
|
chr4_+_101180625
|
17.882
|
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr10_+_69488049
|
17.857
|
|
Ank3
|
ankyrin 3, epithelial
|
|
chr9_-_91255831
|
17.552
|
|
Zic1
|
zinc finger protein of the cerebellum 1
|
|
chr5_+_89012596
|
17.420
|
NM_027530
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr13_+_42775991
|
17.223
|
NM_198419
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr12_+_30213224
|
16.617
|
NM_001093775
NM_001093778
NM_008666
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr13_+_46597271
|
16.606
|
NM_026056
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast)
|
|
chr1_-_42724504
|
15.463
|
|
2610017I09Rik
|
RIKEN cDNA 2610017I09 gene
|
|
chr14_+_28689289
|
15.444
|
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr13_+_58909254
|
15.088
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr13_+_42776161
|
14.886
|
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr7_-_20284370
|
14.833
|
|
Apoe
|
apolipoprotein E
|
|
chr10_-_63552994
|
14.816
|
NM_178678
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr3_+_68376319
|
14.414
|
|
Schip1
|
schwannomin interacting protein 1
|
|
chr5_+_89012537
|
14.200
|
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr5_+_89012547
|
14.164
|
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr7_-_20284500
|
14.047
|
NM_009696
|
Apoe
|
apolipoprotein E
|
|
chr14_-_109313298
|
13.931
|
|
Slitrk1
|
SLIT and NTRK-like family, member 1
|
|
chr2_+_97306600
|
13.303
|
|
|
|
|
chr3_+_67696141
|
13.220
|
NM_001113419
NM_177585
|
Iqcj-schip1
Iqcj
|
IQ motif containing J-schwannomin interacting protein 1 read-through transcript
IQ motif containing J
|
|
chr5_+_90889913
|
13.205
|
NM_009654
|
Alb
|
albumin
|
|
chrX_-_35605544
|
12.861
|
|
Fam70a
|
family with sequence similarity 70, member A
|
|
chr3_+_68376289
|
12.848
|
NM_013928
|
Schip1
|
schwannomin interacting protein 1
|
|
chr5_+_90889937
|
12.700
|
|
Alb
|
albumin
|
|
chr14_-_109313333
|
12.396
|
|
Slitrk1
|
SLIT and NTRK-like family, member 1
|
|
chr14_-_109313447
|
12.359
|
NM_199065
|
Slitrk1
|
SLIT and NTRK-like family, member 1
|
|
chr15_+_4325490
|
12.219
|
NM_177355
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr18_+_61123204
|
12.050
|
NM_009792
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr18_+_37925233
|
11.968
|
NM_033595
|
Pcdhga12
|
protocadherin gamma subfamily A, 12
|
|
chr16_-_42340613
|
11.726
|
NM_008083
|
Gap43
|
growth associated protein 43
|
|
chr18_+_37834003
|
11.693
|
|
Pcdhga3
|
protocadherin gamma subfamily A, 3
|
|
chr7_+_31092787
|
11.547
|
|
Clip3
|
CAP-GLY domain containing linker protein 3
|
|
chr2_+_61884596
|
11.544
|
NM_033552
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10
|
|
chr13_+_58909236
|
11.479
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr6_-_13785233
|
11.468
|
|
Gpr85
|
G protein-coupled receptor 85
|
|
chrX_+_155897136
|
11.201
|
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr13_+_58909168
|
11.136
|
NM_001025074
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr3_+_114783882
|
11.111
|
NM_153157
|
Olfm3
|
olfactomedin 3
|
|
chr10_-_31410391
|
11.046
|
|
Nkain2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr8_+_12984287
|
10.696
|
|
Mcf2l
|
mcf.2 transforming sequence-like
|
|
chr4_+_101223198
|
10.614
|
NM_001164583
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr9_-_73781373
|
10.570
|
NM_001081153
|
Unc13c
|
unc-13 homolog C (C. elegans)
|
|
chr8_+_12949417
|
10.538
|
NM_001159485
NM_001159486
|
Mcf2l
|
mcf.2 transforming sequence-like
|
|
chr18_-_43219062
|
10.348
|
NM_028392
|
Ppp2r2b
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform
|
|
chr4_-_60594959
|
10.323
|
NM_001122647
|
Mup10
LOC100048884
Mup1
|
major urinary protein 10
novel member of the major urinary protein (Mup) gene family
major urinary protein 1
|
|
chr9_-_49607164
|
10.172
|
NM_001081445
NM_001113204
NM_010875
|
Ncam1
|
neural cell adhesion molecule 1
|
|
chr12_+_110783593
|
10.008
|
|
Meg3
|
maternally expressed 3
|
|
chr6_-_102414660
|
9.979
|
NM_008779
|
Cntn3
|
contactin 3
|
|
chr6_-_92484838
|
9.963
|
NM_001134459
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr2_+_97307652
|
9.650
|
NM_178725
|
Lrrc4c
|
leucine rich repeat containing 4C
|
|
chr12_+_90191833
|
9.585
|
NM_172544
|
Nrxn3
|
neurexin III
|
|
chr8_-_87461251
|
9.304
|
NM_019945
|
Mast1
|
microtubule associated serine/threonine kinase 1
|
|
chr3_+_17695743
|
9.259
|
|
6430547I21Rik
|
RIKEN cDNA 6430547I21 gene
|
|
chr5_-_84846256
|
9.156
|
|
Epha5
|
Eph receptor A5
|
|
chr11_+_98629709
|
9.132
|
|
|
|
|
chr11_+_87394647
|
9.129
|
|
Sept4
|
septin 4
|
|
chr18_-_32109233
|
9.092
|
NM_001025381
|
Gpr17
|
G protein-coupled receptor 17
|
|
chr13_+_58909632
|
9.070
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr10_+_126857922
|
9.038
|
NM_001168293
NM_027900
|
R3hdm2
|
R3H domain containing 2
|
|
chr4_+_138012860
|
9.026
|
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr11_+_29607599
|
9.015
|
|
Rtn4
|
reticulon 4
|
|
chr4_-_60889249
|
8.950
|
NM_001134674
|
Mup13
Mup19
Mup1
|
major urinary protein 13
major urinary protein 19
major urinary protein 1
|
|
chr6_+_104443037
|
8.906
|
NM_017383
|
Cntn6
|
contactin 6
|
|
chr12_+_39506844
|
8.897
|
NM_007960
|
Etv1
|
ets variant gene 1
|
|
chr2_-_25325248
|
8.831
|
NM_008963
|
Ptgds
|
prostaglandin D2 synthase (brain)
|
|
chr3_-_146432922
|
8.816
|
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr16_+_96689206
|
8.694
|
NM_008791
|
Pcp4
|
Purkinje cell protein 4
|
|
chr12_+_110806715
|
8.687
|
|
Meg3
|
maternally expressed 3
|
|
chr2_-_25325196
|
8.606
|
|
Ptgds
|
prostaglandin D2 synthase (brain)
|
|
chr15_-_8660729
|
8.601
|
NM_148938
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr19_+_27291897
|
8.581
|
|
Vldlr
|
very low density lipoprotein receptor
|
|
chr12_+_101447413
|
8.482
|
|
Calm1
|
calmodulin 1
|
|
chr18_+_37925619
|
8.410
|
|
Pcdhga12
|
protocadherin gamma subfamily A, 12
|
|
chr9_-_83003967
|
8.406
|
|
|
|
|
chr16_+_96689373
|
8.197
|
|
Pcp4
|
Purkinje cell protein 4
|
|
chr12_+_118896501
|
8.158
|
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr4_-_109960078
|
8.156
|
NM_001038698
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chrX_-_40627962
|
8.128
|
NM_011855
|
Odz1
|
odd Oz/ten-m homolog 1 (Drosophila)
|
|
chrX_-_35605626
|
8.105
|
NM_172930
|
Fam70a
|
family with sequence similarity 70, member A
|
|
chrX_+_155897056
|
8.082
|
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr15_-_58614642
|
7.976
|
|
Tmem65
|
transmembrane protein 65
|
|
chr2_-_77357674
|
7.939
|
NM_001113400
|
Zfp385b
|
zinc finger protein 385B
|
|
chr18_-_31476721
|
7.912
|
NM_009065
|
Rit2
|
Ras-like without CAAX 2
|
|
chr12_+_101447480
|
7.870
|
|
Calm1
|
calmodulin 1
|
|
chrX_+_10173557
|
7.727
|
|
|
|
|
chr3_-_146444377
|
7.674
|
NM_001164200
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr3_-_117063793
|
7.574
|
NM_177664
|
D3Bwg0562e
|
DNA segment, Chr 3, Brigham & Women's Genetics 0562 expressed
|
|
chr1_-_182123596
|
7.548
|
NM_001163290
|
Adck3
|
aarF domain containing kinase 3
|
|
chrX_-_100381973
|
7.449
|
NM_008671
|
Nap1l2
|
nucleosome assembly protein 1-like 2
|
|
chr18_+_37849488
|
7.410
|
NM_033575
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2
|
|
chr11_+_87394543
|
7.398
|
|
Sept4
|
septin 4
|
|
chr8_+_116729508
|
7.374
|
NM_173016
|
Vat1l
|
vesicle amine transport protein 1 homolog-like (T. californica)
|
|
chr18_+_37979242
|
7.360
|
|
Pcdhgc5
|
protocadherin gamma subfamily C, 5
|
|
chr5_-_121459496
|
7.217
|
NM_011286
|
Rph3a
|
rabphilin 3A
|
|
chr3_-_80606500
|
7.171
|
|
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2)
|
|
chr9_-_112134984
|
7.117
|
NM_001177616
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21
|
|
chr4_-_86876328
|
7.112
|
NM_172426
|
Slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr5_+_90890764
|
7.074
|
|
Alb
|
albumin
|
|
chr18_+_37595274
|
7.069
|
NM_053137
|
Pcdhb12
|
protocadherin beta 12
|
|
chr14_-_104213981
|
7.045
|
|
|
|
|
chr11_+_87394609
|
7.022
|
NM_011129
|
Sept4
|
septin 4
|
|
chr15_-_73640331
|
6.976
|
|
Ndufb4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4
|
|
chr10_+_5151833
|
6.950
|
NM_001079686
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr9_+_21742717
|
6.948
|
NM_144935
|
BC018242
|
cDNA sequence BC018242
|
|
chr13_+_20277293
|
6.826
|
NM_080288
|
Elmo1
|
engulfment and cell motility 1, ced-12 homolog (C. elegans)
|
|
chr1_-_5060365
|
6.799
|
NM_001177795
|
Rgs20
|
regulator of G-protein signaling 20
|
|
chr8_-_87461220
|
6.740
|
|
Mast1
|
microtubule associated serine/threonine kinase 1
|
|
chr16_-_84955488
|
6.716
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr10_+_59763520
|
6.695
|
|
Psap
|
prosaposin
|
|
chr12_-_32398730
|
6.631
|
NM_175191
|
Gpr22
|
G protein-coupled receptor 22
|
|
chr12_+_109843975
|
6.620
|
NM_001163395
|
Evl
|
Ena-vasodilator stimulated phosphoprotein
|
|
chr19_-_55177562
|
6.608
|
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr16_-_84955086
|
6.542
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr6_+_77193078
|
6.532
|
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr11_-_79317471
|
6.509
|
|
Omg
|
oligodendrocyte myelin glycoprotein
|
|
chr18_+_37979199
|
6.509
|
NM_033583
|
Pcdhgb4
Pcdhgc5
|
protocadherin gamma subfamily B, 4
protocadherin gamma subfamily C, 5
|
|
chr1_+_34364633
|
6.482
|
|
Dst
|
dystonin
|
|
chr15_+_91958558
|
6.420
|
NM_007727
|
Cntn1
|
contactin 1
|
|
chr14_+_65885454
|
6.408
|
NM_015795
|
Fbxo16
|
F-box protein 16
|
|
chr12_+_50484907
|
6.368
|
|
Foxg1
|
forkhead box G1
|
|
chr19_+_8915018
|
6.213
|
|
Bscl2
|
Bernardinelli-Seip congenital lipodystrophy 2 homolog (human)
|
|
chrX_+_132424640
|
6.197
|
|
Bhlhb9
|
basic helix-loop-helix domain containing, class B9
|
|
chr2_+_155215225
|
6.162
|
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2
|
|
chr14_+_66963139
|
6.150
|
NM_019675
|
Stmn4
|
stathmin-like 4
|
|
chr15_-_37389073
|
6.079
|
NM_001170867
NM_001170868
|
Ncald
|
neurocalcin delta
|
|
chr8_+_115093942
|
6.075
|
NM_130457
|
Cntnap4
|
contactin associated protein-like 4
|
|
chr4_+_101180580
|
6.046
|
NM_001164584
NM_001164585
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr14_-_31304341
|
6.000
|
NM_028981
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr18_-_31606908
|
5.981
|
NM_009308
|
Syt4
|
synaptotagmin IV
|
|
chr3_+_136598184
|
5.975
|
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform
|
|
chr16_+_96689322
|
5.837
|
|
|
|
|
chr19_+_56798916
|
5.800
|
|
Adrb1
|
adrenergic receptor, beta 1
|
|
chr6_+_115189004
|
5.787
|
|
Syn2
|
synapsin II
|
|
chr6_+_77192839
|
5.771
|
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr2_-_73498303
|
5.754
|
NM_001166604
NM_175752
|
Chn1
|
chimerin (chimaerin) 1
|
|
chr12_+_110800212
|
5.751
|
|
Meg3
|
maternally expressed 3
|
|
chr10_+_84218792
|
5.714
|
NM_001024918
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr2_+_59192308
|
5.697
|
|
Pkp4
|
plakophilin 4
|
|
chr18_-_42900793
|
5.678
|
|
Ppp2r2b
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform
|
|
chr5_-_139606339
|
5.678
|
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta
|
|
chr6_+_124948627
|
5.621
|
NM_001145926
|
C530028O21Rik
|
RIKEN cDNA C530028O21 gene
|
|
chr14_-_55613384
|
5.569
|
NM_080728
|
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta
|
|
chr7_+_148817700
|
5.558
|
|
Ap2a2
|
adaptor protein complex AP-2, alpha 2 subunit
|
|
chr2_-_148558110
|
5.537
|
NM_019632
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta
|
|
chr3_-_19595395
|
5.527
|
NM_205769
|
Crh
|
corticotropin releasing hormone
|
|
chr6_+_134932018
|
5.522
|
NM_001109914
|
Apold1
|
apolipoprotein L domain containing 1
|
|
chrX_+_20265093
|
5.499
|
|
Cdk16
|
cyclin-dependent kinase 16
|
|
chr7_-_31840647
|
5.430
|
NM_019503
NM_052992
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1
|
|
chr10_+_98486068
|
5.409
|
|
Atp2b1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chrX_+_139997806
|
5.361
|
NM_008778
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr13_+_83803830
|
5.352
|
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chr14_-_80171003
|
5.348
|
NM_001042726
NM_021543
|
Pcdh8
|
protocadherin 8
|
|
chrX_+_133371611
|
5.321
|
|
Plp1
|
proteolipid protein (myelin) 1
|
|
chr2_-_23904343
|
5.270
|
|
Hnmt
|
histamine N-methyltransferase
|
|
chr5_+_137506122
|
5.270
|
NM_001039385
|
Vgf
|
VGF nerve growth factor inducible
|
|
chr17_-_43803963
|
5.258
|
NM_028711
|
Slc25a27
|
solute carrier family 25, member 27
|
|
chrX_-_100381703
|
5.252
|
|
Nap1l2
|
nucleosome assembly protein 1-like 2
|
|
chr9_-_107571294
|
5.110
|
|
Slc38a3
|
solute carrier family 38, member 3
|
|
chr4_-_136441982
|
5.100
|
NM_009777
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide
|
|
chr12_+_39510297
|
5.095
|
NM_001163154
|
Etv1
|
ets variant gene 1
|
|
chr8_+_56122680
|
5.066
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr3_-_84024766
|
5.058
|
NM_030706
|
Trim2
|
tripartite motif-containing 2
|
|
chr12_+_110783786
|
5.012
|
|
Meg3
|
maternally expressed 3
|
|
chr2_+_20211539
|
5.004
|
NM_001081006
|
Etl4
|
enhancer trap locus 4
|
|
chr18_+_37460452
|
4.987
|
NM_053128
|
Pcdhb3
|
protocadherin beta 3
|
|
chr19_-_7500486
|
4.973
|
|
|
|
|
chr12_-_4344470
|
4.970
|
|
Ncoa1
|
nuclear receptor coactivator 1
|
|
chr6_-_92431385
|
4.970
|
NM_001134460
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr19_+_27291507
|
4.960
|
NM_001161420
NM_013703
|
Vldlr
|
very low density lipoprotein receptor
|
|
chr7_-_67285104
|
4.937
|
NM_001082962
|
Snrpn
|
small nuclear ribonucleoprotein N
|
|
chr1_-_67085404
|
4.927
|
NM_001190984
NM_001190985
NM_021295
|
Lancl1
|
LanC (bacterial lantibiotic synthetase component C)-like 1
|
|
chr7_-_31840594
|
4.901
|
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1
|
|
chr6_+_58783693
|
4.876
|
NM_028705
|
Herc3
|
hect domain and RLD 3
|
|
chr5_+_89012600
|
4.873
|
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chrM_+_12383
|
4.852
|
|
|
|
|
chrX_-_161764912
|
4.851
|
NM_183427
|
Glra2
|
glycine receptor, alpha 2 subunit
|
|
chr5_-_115636064
|
4.835
|
NM_013879
|
Cabp1
|
calcium binding protein 1
|
|
chr9_-_107571304
|
4.799
|
|
Slc38a3
|
solute carrier family 38, member 3
|
|
chr1_+_169548381
|
4.771
|
NM_001159731
|
Rxrg
|
retinoid X receptor gamma
|
|
chr9_-_55983967
|
4.715
|
|
Tspan3
|
tetraspanin 3
|
|
chr8_+_114062242
|
4.642
|
NM_001168623
|
Znrf1
|
zinc and ring finger 1
|
|
chr9_-_60535274
|
4.641
|
NM_001146047
|
Lrrc49
|
leucine rich repeat containing 49
|
|
chr3_-_80606704
|
4.628
|
NM_001039195
NM_001083806
NM_013540
|
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2)
|
|
chr5_+_107926763
|
4.608
|
NM_001007574
NM_001168557
|
A830010M20Rik
|
RIKEN cDNA A830010M20 gene
|
|
chr6_+_8900018
|
4.602
|
NM_008751
|
Nxph1
|
neurexophilin 1
|